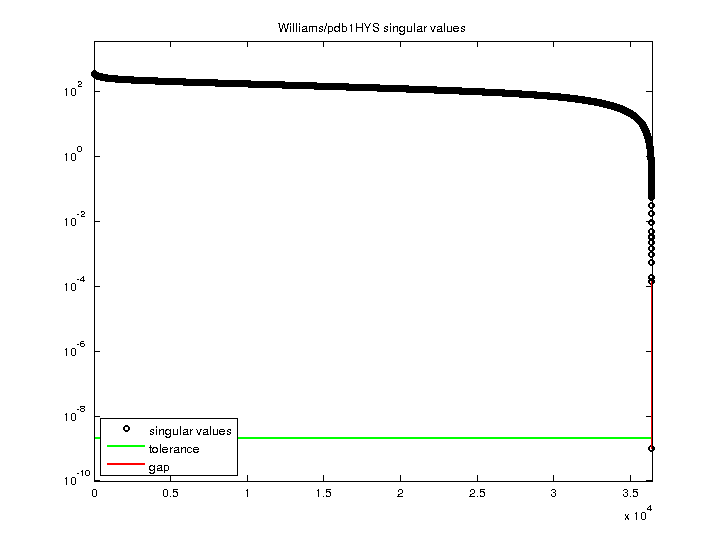
Williams/pdb1HYS
Protein: protein data bank 1HYS. Williams et al.
| Name |
pdb1HYS |
| Group |
Williams |
| Matrix ID |
2373 |
|
Num Rows
|
36,417 |
|
Num Cols
|
36,417 |
|
Nonzeros
|
4,344,765 |
|
Pattern Entries
|
4,344,765 |
|
Kind
|
Weighted Undirected Graph |
|
Symmetric
|
Yes |
|
Date
|
2008 |
|
Author
|
S. G. Sarafianos et al |
|
Editor
|
S. Williams, L. Oliker, R. Vuduc, J. Shalf, K. Yelick, J. Demmel |
| Structural Rank |
|
| Structural Rank Full |
|
|
Num Dmperm Blocks
|
|
|
Strongly Connect Components
|
1 |
|
Num Explicit Zeros
|
0 |
|
Pattern Symmetry
|
100% |
|
Numeric Symmetry
|
100% |
|
Cholesky Candidate
|
yes |
|
Positive Definite
|
yes |
|
Type
|
real |
| SVD Statistics |
| Matrix Norm |
3.523937e+02 |
| Minimum Singular Value |
9.970386e-10 |
| Condition Number |
3.534404e+11
|
| Rank |
36,411 |
| sprank(A)-rank(A) |
|
| Null Space Dimension |
6 |
| Full Numerical Rank? |
no |
| Download Singular Values |
MATLAB
|
| Download |
MATLAB
Rutherford Boeing
Matrix Market
|
| Notes |
Matrices used by S. Williams et al for sparse matrix multiplication on GPUs.
14 matrices were used in the following paper:
S. Williams, L. Oliker, R. Vuduc, J. Shalf, K. Yelick, J. Demmel,
"Optimization of Sparse Matrix-Vector Multiplication on Emerging Multicore
Platforms", Parallel Computing Volume 35, Issue 3, March 2009, Pages
178-194. Special issue on Revolutionary Technologies for Acceleration of
Emerging Petascale Applications.
https://hpcrd.lbl.gov/~swwilliams/research/papers/parco08_spmv.pdf
http://dx.doi.org/10.1016/j.parco.2008.12.006
This same set of 14 matrices was also used in a subsequent technical report by
NVIDIA:
http://www.nvidia.com/object/nvidia_research_pub_001.html "Efficient Sparse
Matrix-Vector Multiplication on CUDA" Nathan Bell and Michael Garland, in,
"NVIDIA Technical Report NVR-2008-004", December 2008
file Name dim* nnz description
dense2 Dense 2K 4.0M dense matrix in sparse format
pdb1HYS Protein 36K 4.3M protein data bank 1HYS
consph FEM/Spheres 83K 6.0M FEM concentric spheres
cant FEM/Cantilever 62K 4.0M FEM cantilever
pwtk Wind Tunnel 218K 11.6M pressurized wind tunnel
rma10 FEM/Harbor 47K 2.37M 3D CFD of Charleston Harbor
qcd5_4 QCD 49K 1.90M quark propagators (QCD/LGT)
shipsec1 FEM/Ship 141K 3.98M FEM Ship section / detail
mac_econ_fwd500 Economics 207K 1.27M Macroeconomic model
mc2depi Epidemiology 526K 2.1M 2D Markov model of epidemic
cop20k_A FEM/Accelerator 121K 2.62M Accelerator cavity design
scircuit Circuit 171K 959K Motorola circuit simulation
webbase-1M webbase 1M 3.1M Web connectivity matrix
rail4284 LP 4Kx1.1M 11.3M Railways set cover,
constraint matrix
(*) the matrix is square if only one dimension listed.
Six of the matrices are nearly identical to the matrices already in the
UF Collection. They are thus not included in the UF Collection. See
the README.txt file for this collection for details.
I presume the pdb1HYS matrix comes from this source:
http://www.rcsb.org/pdb/explore.do?structureId=1HYS
http://dx.doi.org/10.2210/pdb1hys/pdb
Crystal structure of HIV-1 reverse transcriptase in complex with a
polypurine tract RNA:DNA.
Sarafianos, S.G., Das, K., Tantillo, C., Clark Jr., A.D., Ding,
J., Whitcomb, J.M., Boyer, P.L., Hughes, S.H., Arnold, E.
Journal: (2001) EMBO J. 20: 1449-1461
PubMed: 11250910
PubMedCentral: PMC145536
DOI: 10.1093/emboj/20.6.1449
Search Related Articles in PubMed
PubMed Abstract:
We have determined the 3.0 A resolution structure of wild-type HIV-1
reverse transcriptase in complex with an RNA:DNA oligonucleotide whose
sequence includes a purine-rich segment from the HIV-1 genome called the
polypurine tract (PPT). The PPT is resistant to ribonuclease... [ Read
More & Search PubMed Abstracts ] We have determined the 3.0 A resolution
structure of wild-type HIV-1 reverse transcriptase in complex with an
RNA:DNA oligonucleotide whose sequence includes a purine-rich segment from
the HIV-1 genome called the polypurine tract (PPT). The PPT is resistant
to ribonuclease H (RNase H) cleavage and is used as a primer for second
DNA strand synthesis. The "RNase H primer grip", consisting of amino
acids that interact with the DNA primer strand, may contribute to RNase H
catalysis and cleavage specificity. Cleavage specificity is also
controlled by the width of the minor groove and the trajectory of the
RNA:DNA, both of which are sequence dependent. An unusual "unzipping" of 7
bp occurs in the adenine stretch of the PPT: an unpaired base on the
template strand takes the base pairing out of register and then, following
two offset base pairs, an unpaired base on the primer strand
re-establishes the normal register. The structural aberration extends to
the RNase H active site and may play a role in the resistance of PPT to
RNase H cleavage.
|