Schulthess/N_pid
biochemical network; left nullspace is required
| Name |
N_pid |
| Group |
Schulthess |
| Matrix ID |
2553 |
|
Num Rows
|
3,625 |
|
Num Cols
|
3,923 |
|
Nonzeros
|
8,054 |
|
Pattern Entries
|
8,054 |
|
Kind
|
Biochemical Network |
|
Symmetric
|
No |
|
Date
|
2012 |
|
Author
|
P. Schulthess |
|
Editor
|
T. Davis |
| Structural Rank |
2,171 |
| Structural Rank Full |
false |
|
Num Dmperm Blocks
|
640 |
|
Strongly Connect Components
|
1,216 |
|
Num Explicit Zeros
|
0 |
|
Pattern Symmetry
|
0% |
|
Numeric Symmetry
|
0% |
|
Cholesky Candidate
|
no |
|
Positive Definite
|
no |
|
Type
|
integer |
| SVD Statistics |
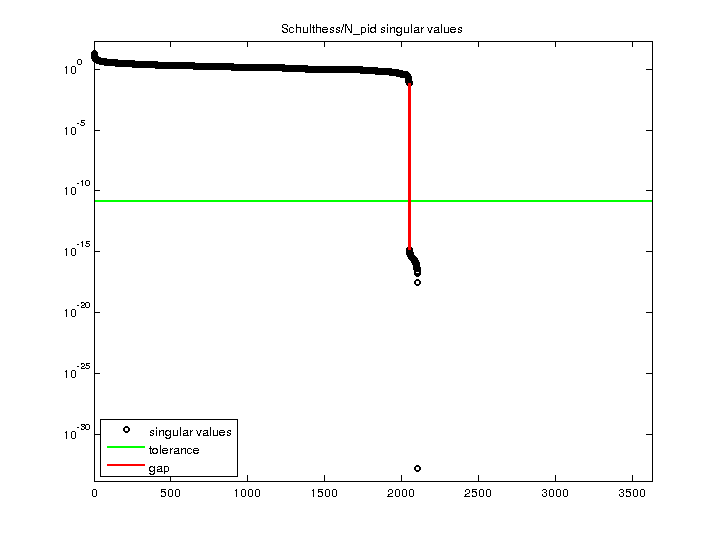
| Matrix Norm |
2.049051e+01 |
| Minimum Singular Value |
0 |
| Condition Number |
Inf
|
| Rank |
2,048 |
| sprank(A)-rank(A) |
123 |
| Null Space Dimension |
1,577 |
| Full Numerical Rank? |
no |
| Download Singular Values |
MATLAB
|
| Download |
MATLAB
Rutherford Boeing
Matrix Market
|
| Notes |
Matrices from Pascal Schulthess, Institute for Pathology,
Chariteplatz 1, 10117 Berlin, Germany.
Three large biochemical networks (N_biocarta, N_pid, and N_reactome).
These are stoichiometric matrices extracted from three biochemical
databases (BioCarta, PID, and REACTOME) describing cell signaling
pathways and protein-protein interaction networks. The goal is to
find the left nullspace of the matrix; in MATLAB notation:
N = null (Problem.A') ;
The matrix (Problem.A')*N will thus be essentially zero.
This can be done much more efficiently with the spqr_rank toolbox by
Leslie Foster and Tim Davis, as:
N = spqr_null (Problem.A') ;
Results:
The matrix A is transposed, then N = null (A) or N = spqr_null (A)
is computed. The size statistic is the memory taken by N.
spqr_null can compute either an explicit matrix N, or an implicit
Householder-based representation. The latter takes less memory.
Matrix: N_biocarta size: 1996 by 1922 (transposed)
spqr_null stats:
flag: 0
rank: 1023
tol: 3.5456e-12
est_sval_upper_bounds: [0.1689 3.4534e-15]
est_sval_lower_bounds: [0.1203 0]
sval_numbers_for_bounds: [1023 1024]
est_norm_A_times_N: 2.4349e-15
spqr_null, implicit: 0.03 sec, norm(A*N) 9e-15 size: 0.08 MB
spqr_null, explicit: 0.10 sec, norm(A*N) 9e-15 size: 0.11 MB
MATLAB null: 3.31 sec, norm(A*N) 2e-13 size: 13.82 MB
all report dim(N) of 899.
Matrix: N_pid size: 3923 by 3625 (transposed)
spqr_null stats:
flag: 0
rank: 2048
tol: 1.3937e-11
est_sval_upper_bounds: [0.0922 5.1310e-15]
est_sval_lower_bounds: [0.0585 0]
sval_numbers_for_bounds: [2048 2049]
est_norm_A_times_N: 1.6751e-15
spqr_null, implicit: 0.05 sec, norm(A*N) 4e-14 size: 0.21 MB
spqr_null, explicit: 0.34 sec, norm(A*N) 4e-14 size: 1.32 MB
MATLAB null: 24.86 sec, norm(A*N) 9e-13 size: 45.73 MB
all report dim(N) of 1577
Matrix: N_reactome size: 16559 by 10204 (transposed)
spqr_null stats:
flag: 0
rank: 9025
tol: 1.1766e-10
est_sval_upper_bounds: [0.6722 1.3042e-14]
est_sval_lower_bounds: [0.0106 0]
sval_numbers_for_bounds: [9025 9026]
est_norm_A_times_N: 9.4695e-15
spqr_null, implicit: 0.95 sec, norm(A*N) 2e-13 size: 7.5 MB
spqr_null, explicit: 3.53 sec, norm(A*N) 2e-13 size: 25.2 MB
MATLAB null: 904.54 sec, norm(A*N) 2e-10 size: 96.2 MB
all report dim(N) of 1179.
|